Through the first half of the 20th century evolution was mostly the science of selection and splitting. To illustrate with diagrams, a simple selection operation looks like this:
...and splitting looks like this:
Then in the 1960s, symbiosis and sex started to look as though they needed another fundamental operation - merging - which looks like this:
These are the basic operations in modern evolutionary theory. As you can see from the diagrams, these basic operations are particulate - in that they operate on discrete, particle-like entities.
With the modern expansion of the domain of evolutionary theory it seems to me that we need to consider some other levels of analysis. In particular I think we should be thinking about graph evolution and tree evolution. I mean "graph" in the sense of graph theory. You could also call it 'network evolution'.
I'm thinking that a node-and-link scheme would be a better match for the evolution of brains, tree branches, circulatory systems, immune systems, respiratory systems - and so on.
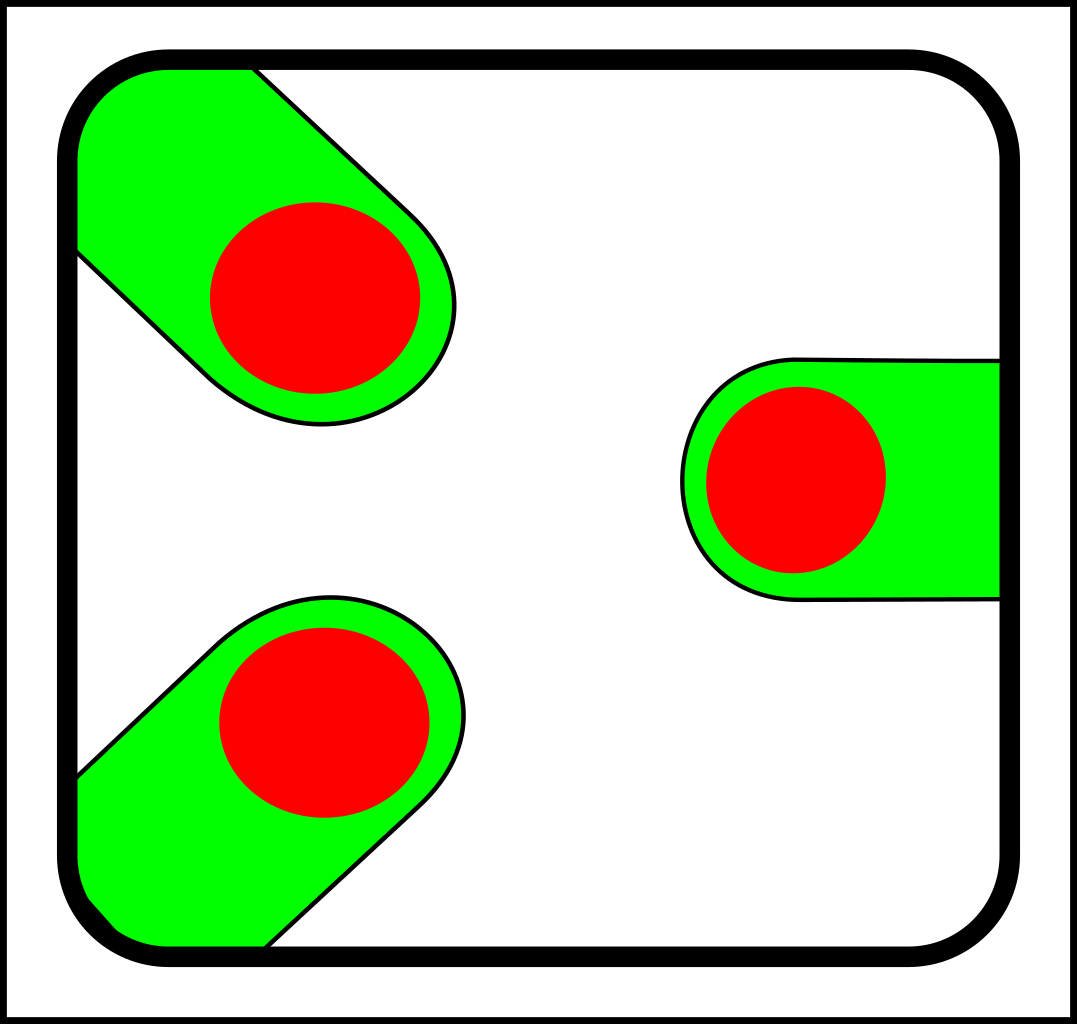
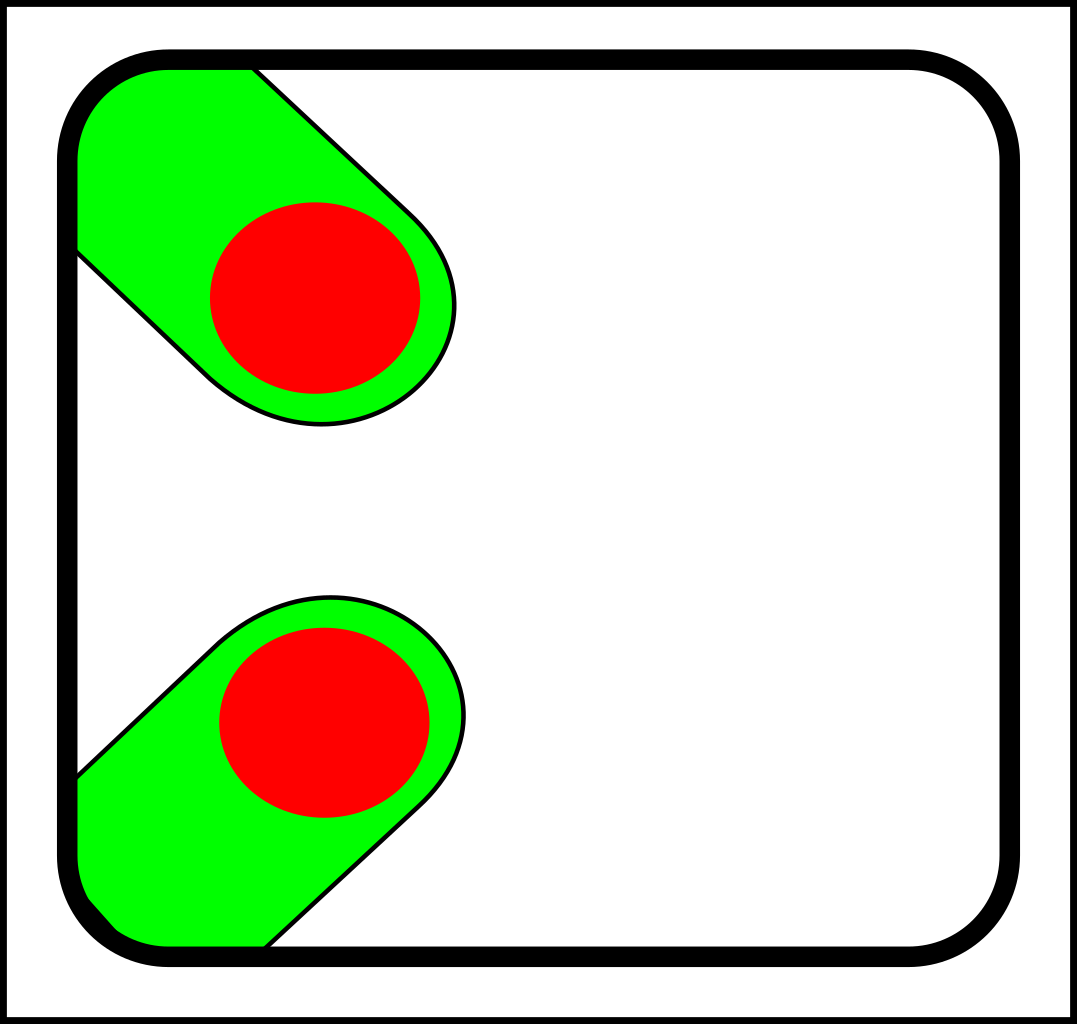
At first glance, this might seem like a case of adding operations that create and destroy links between nodes. That might make sense in computer science, but in nature, links form gradually and a more gradualistic scheme seems more in tune with existing evolutionary theory. Here are the splitting operations:
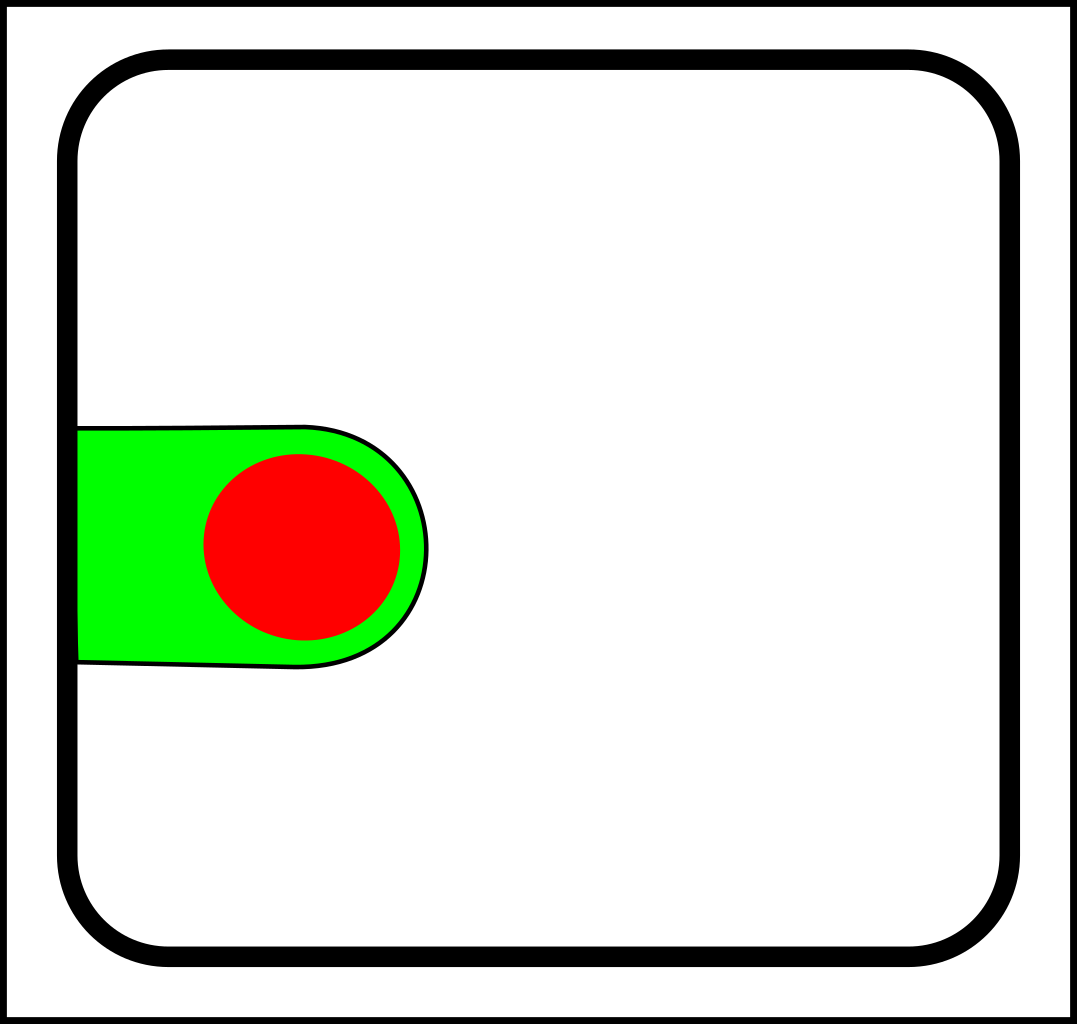 |
-> |
 |
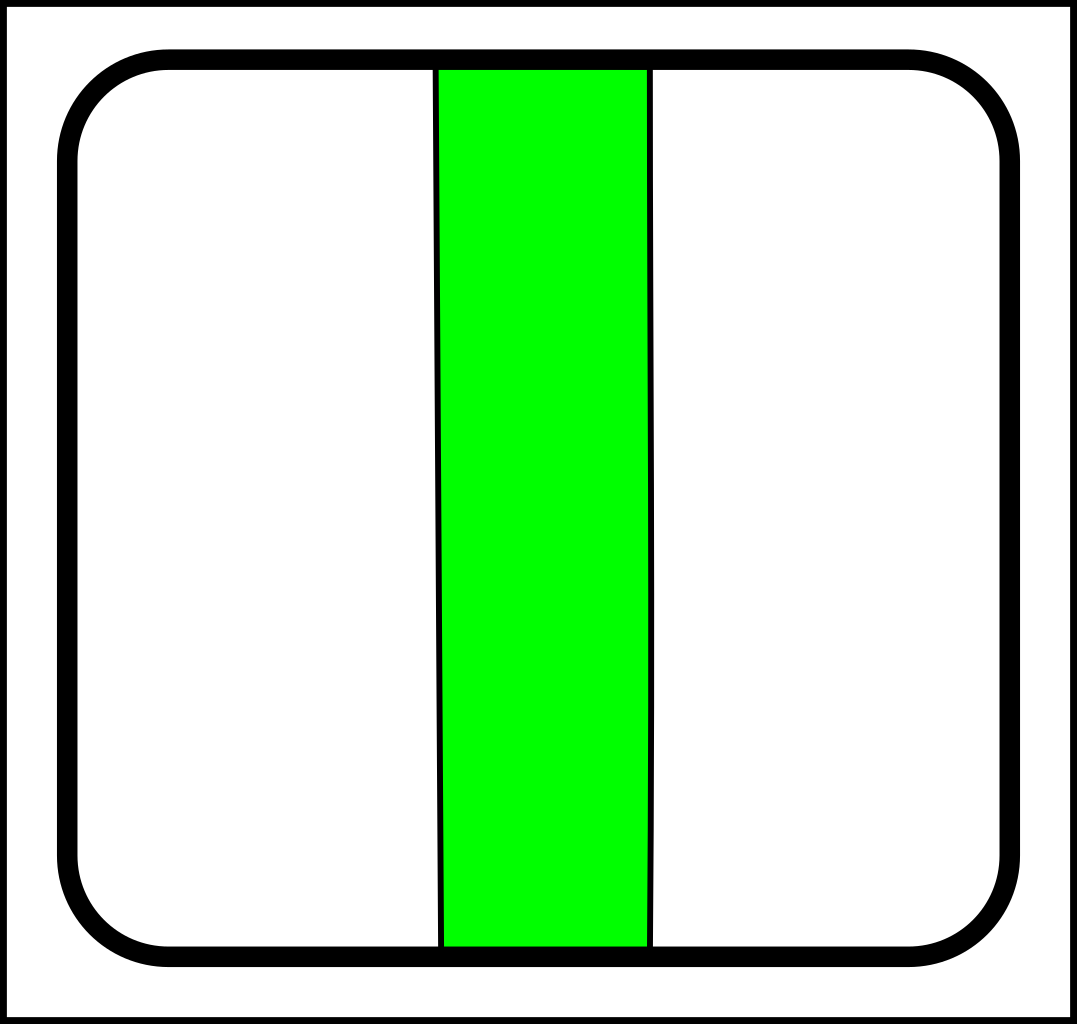 |
-> |
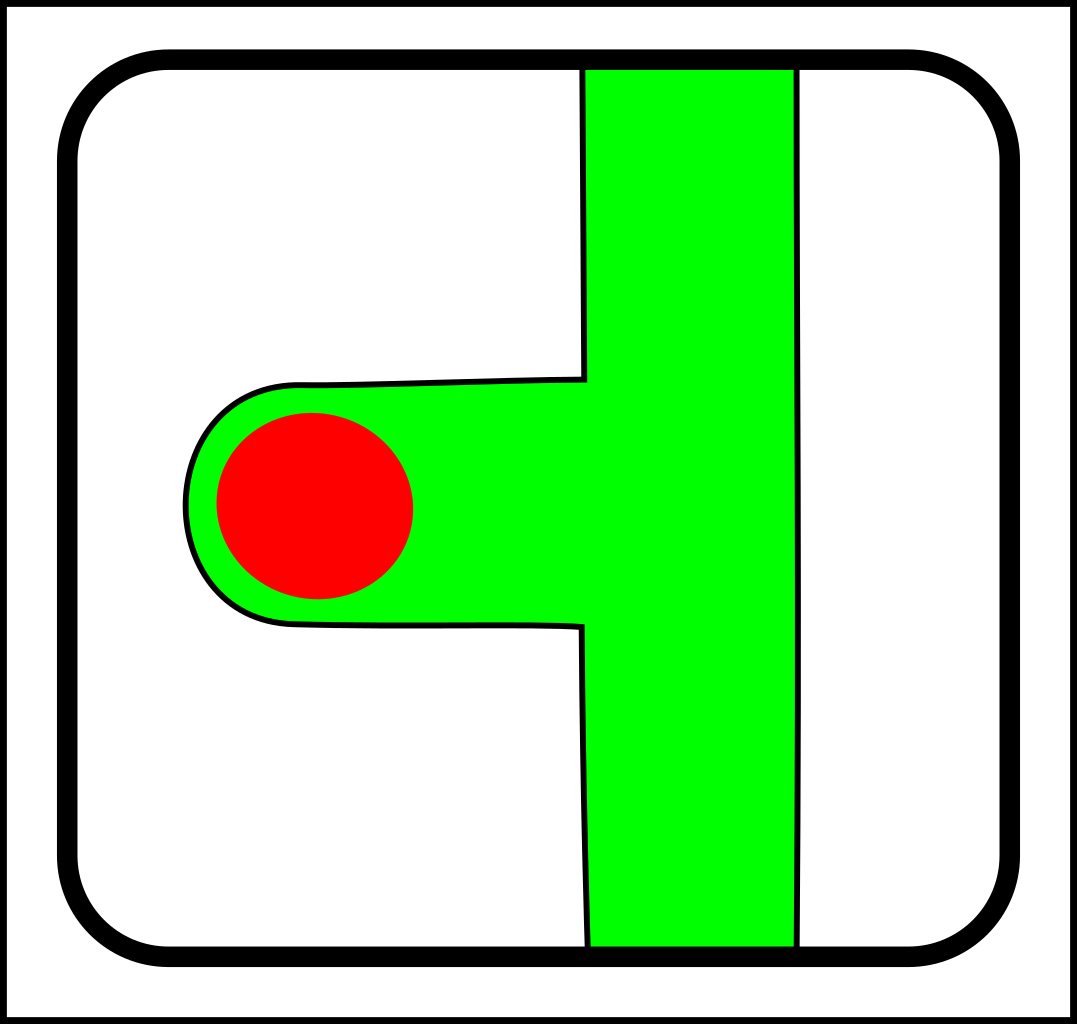 |
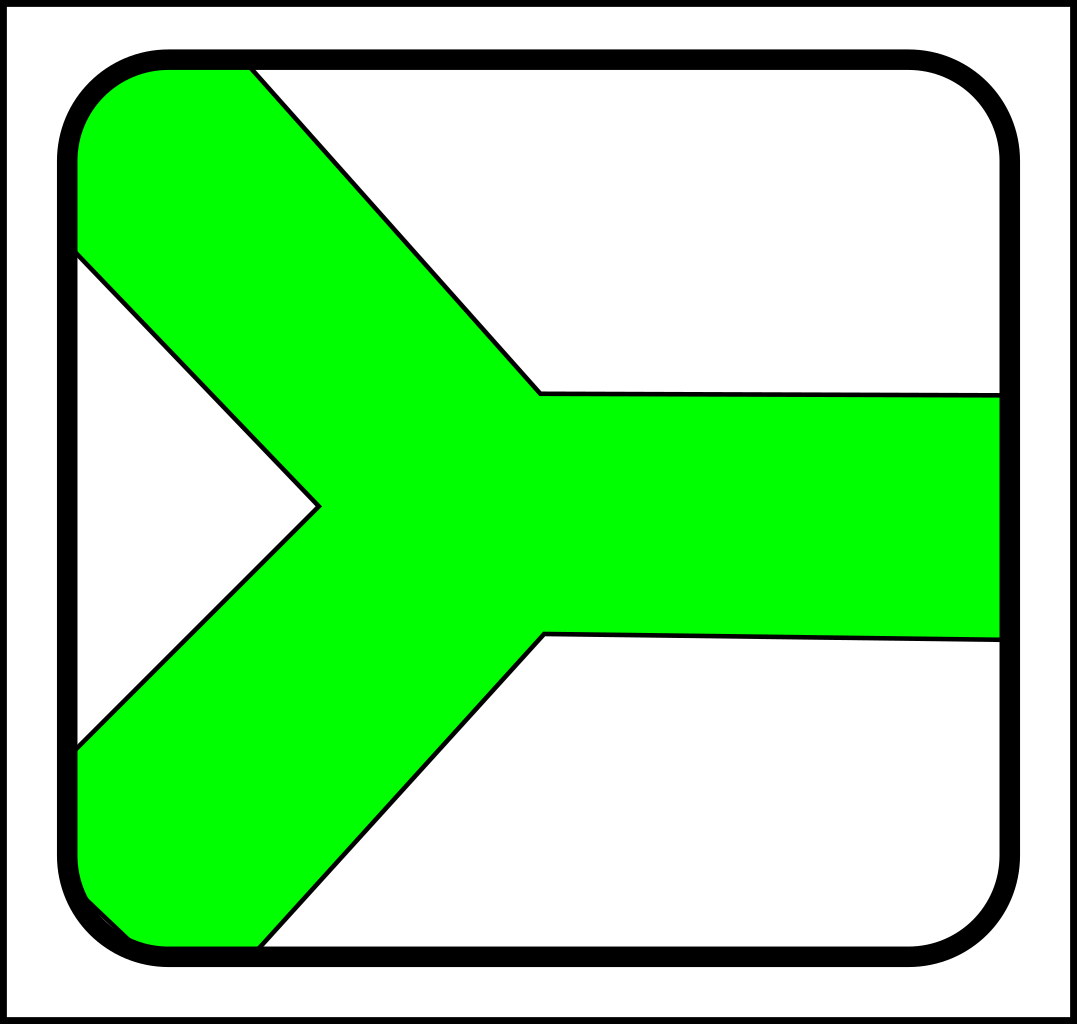 |
-> |
 |
 |
-> |
 |
 |
-> |
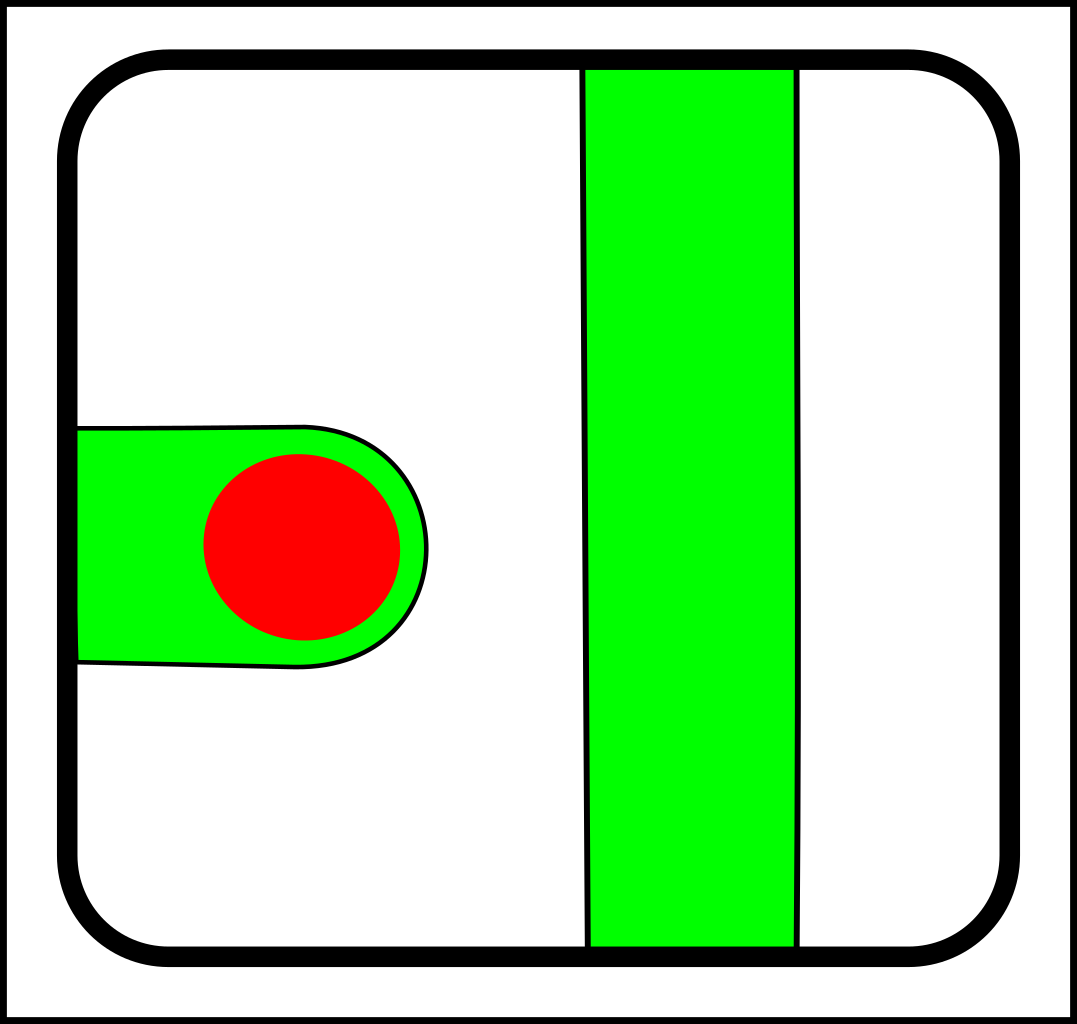 |
 |
-> |
 |
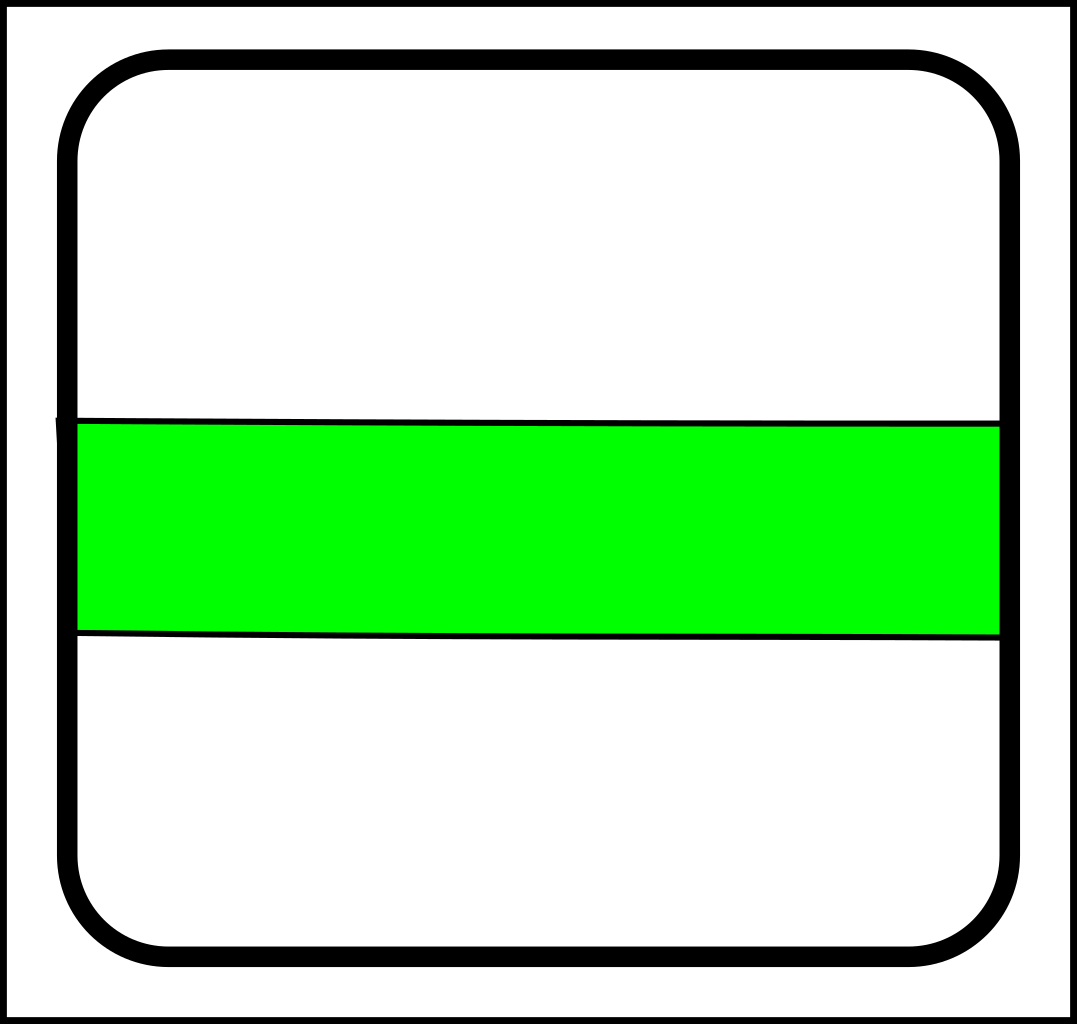
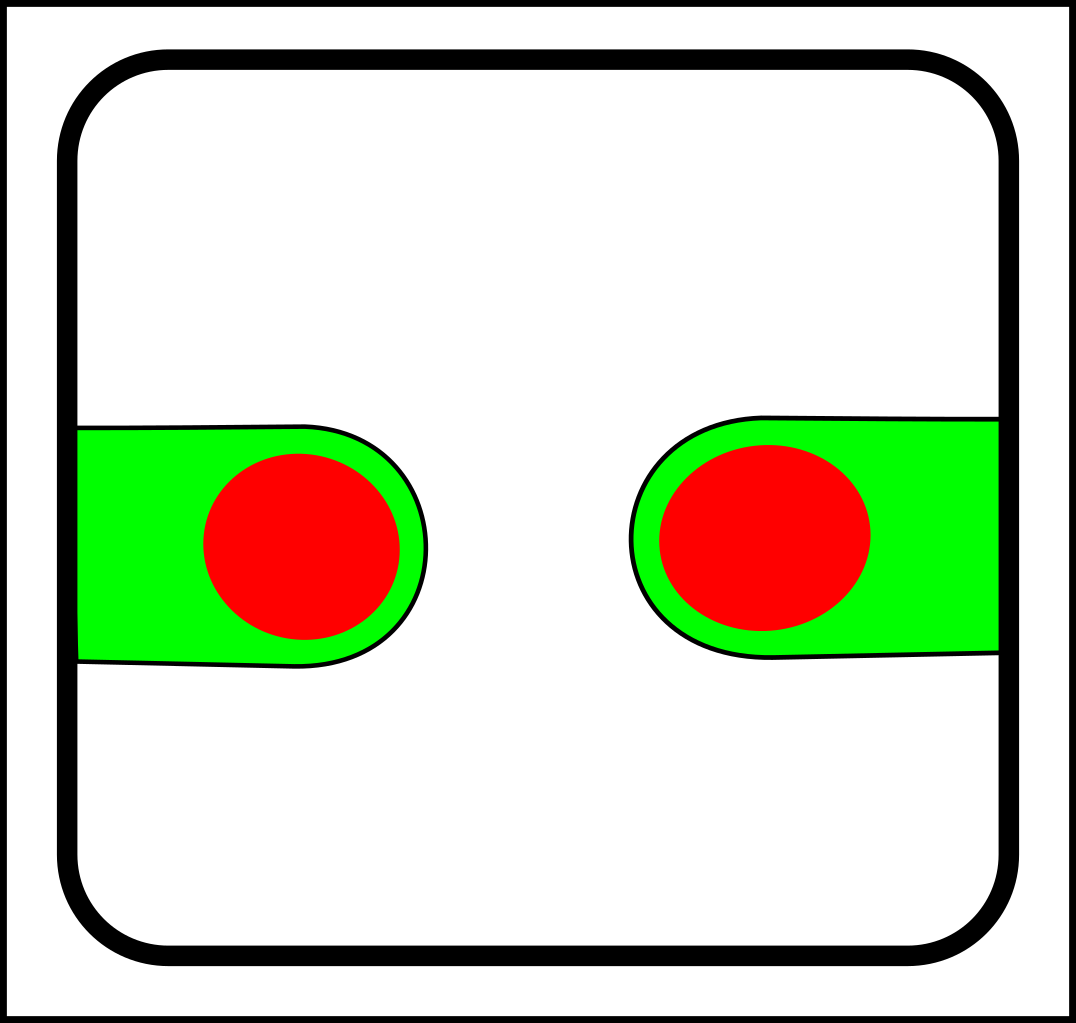
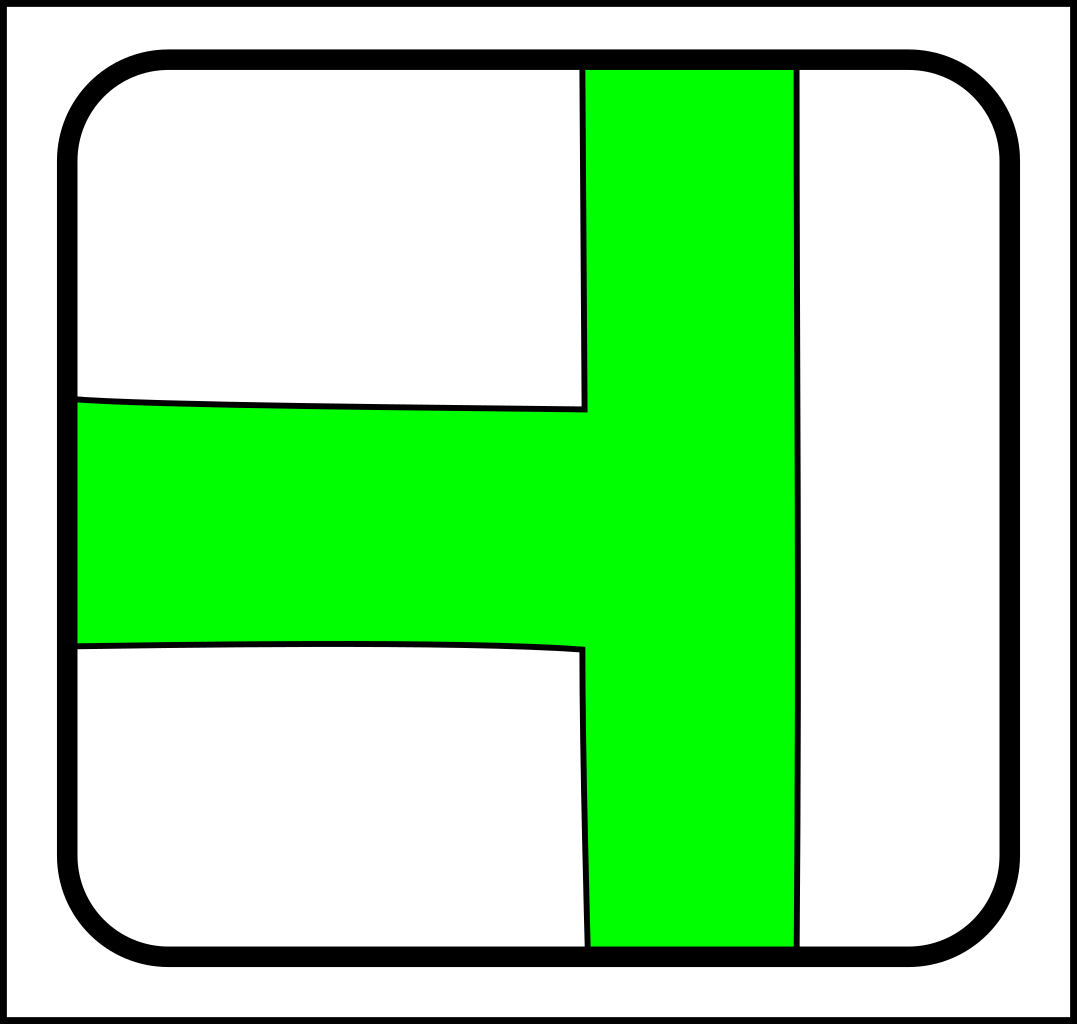
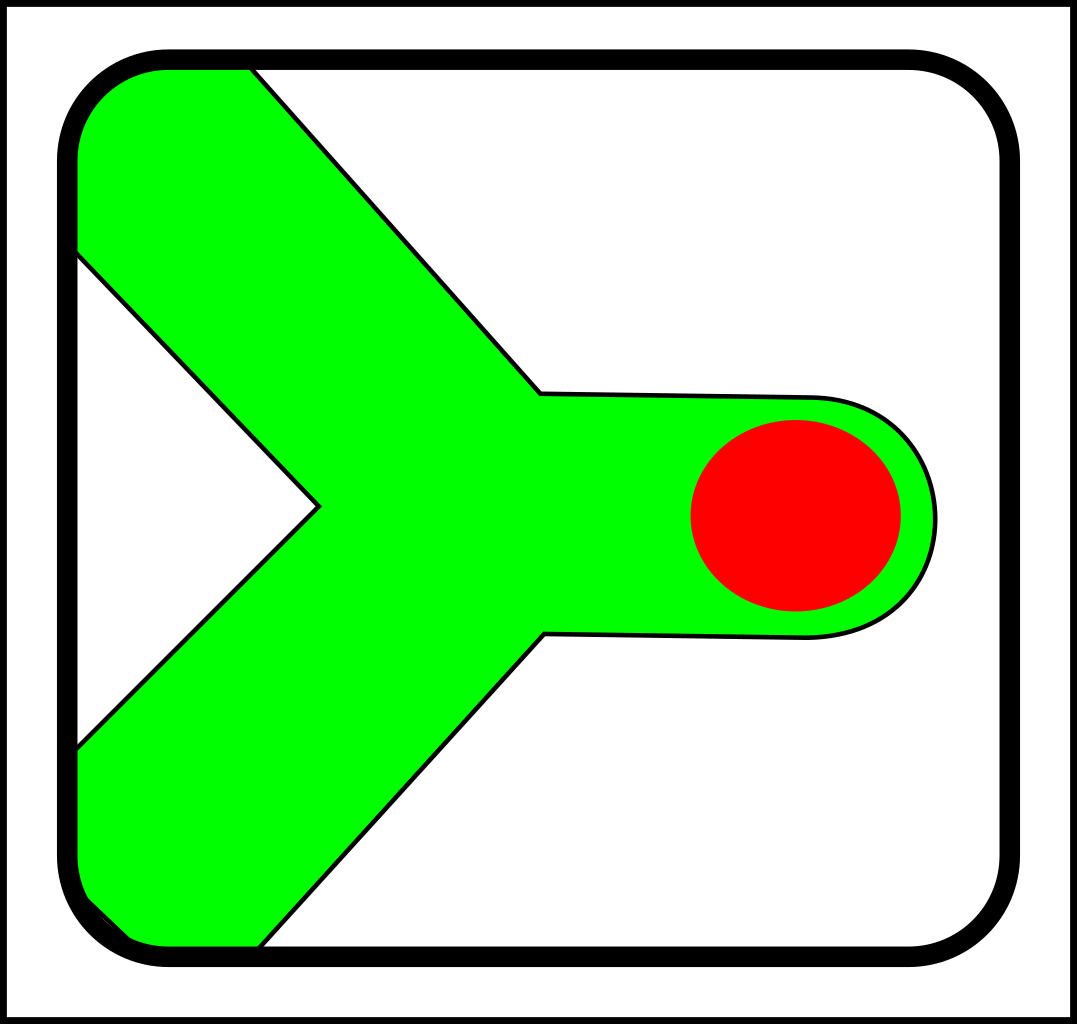
...and here are the corresponding joining operations:
 |
-> |
 |
 |
-> |
 |
 |
-> |
 |
 |
-> |
 |
 |
-> |
 |
 |
-> |
 |
For the sake of simplicity, I've only shown diagrams for nodes with fewer than four links. I've also
omitted selection events - i.e cases where elements (nodes or links) are simply destroyed.
I should probably say a bit more about the motivation for such a scheme. In biology, tree and graph evolution
is often reducible to particle evolution - since the components involved are often composed of particulate cells.
However, this doesn't work so well for brain evolution - since axons and dentrites are so link-like.
It's often possible to model branch tip evolution using conventional Darwinian models which treat branch tips as
individuals. This works quite well in many of the types of inanimate systems considered by Darwinian physics, for example. However this approach works best when flow into the resulting tree-like structures is time-like - i.e. unidirectional and at a constant speed. If resources can flow back out of unpromising roots or newly-shaded branches, conventional Darwinian models based on a population of branch tips do not work so well. Then, more sophisticated models can be useful.
There is a field called evolutionary graph theory -
but that seems to be about something rather different from the topic of this page.



































I'm a member of the Community of Interest on this one (I spend most of my time coding graph-theory software for representing memetic evolution). Obviously you are enumerating a taxonomy of atomic operations - the basic splitting and joining that evolution does. I wouldn't want to get too mathematical (being a computer scientist), but how might these look in G=(V, E) format.
ReplyDeleteI gather the second diagram of splitting might be something like {a1} -> {a1, a2}; which is just a set of nodes.
Where as the fourth, at first glance, might be ({a1}, {}) -> ({a1, a2}, {(a1, a2)}) [but that would produce a "lambda" shape (often seen in cladograms) and not a "Y".] where the links are denoted by the set of edges.
But this doesnt offer a gradualistic scheme (keeping cladists happy) If we took asexual evolution
({i}, {})->({i, j, k}, {(i, j) (i, k)})->({j, k, l, m}, {(j, l), (k, m)})->({k, m}, {}) which produces generational syblings of {j, k} and cousins {k, m} then are you suggesting some kind of distance measure within those sets? eg. distance(k, m)
Hi. If you want an undirected graph, put nodes where three branches meet - and where the branches go off the sides of the diagrams.
DeleteRegarding gradualism, I was thinking that having a link appear between two separate nodes is a dubious operation in many physical systems - and that link formation can be modeled in a more gradual fashion using node splitting and joining operations.